Algorithm Explained¶
from hybridjobs.utility.MoleculeParser import MoleculeData
from hybridjobs.utility.QMUQUBO import QMUQUBO
from hybridjobs.utility.AnnealerOptimizer import Annealer
from hybridjobs.utility.ResultProcess import ResultParser
import time
timestamp = time.strftime("%Y%m%d-%H")
2023-05-27 12:01:05,750 dwave.cloud INFO MainThread Log level for 'dwave.cloud' namespace set to 0
Step 1: Prepare Data¶
In this part, we load the raw molecule data for experiment. The 117 ligand was put in the repository. We assign the relative path to raw_path. The s3_bucket and prefix are used to store the optimization results. We can use the one created with the cloudformation for convenience.
from braket.aws import AwsSession
# initial parameters for experiment data
s3_bucket = AwsSession().default_bucket() # change to the name of bucket created in your deployment
prefix = "molecular-unfolding" # the name of the folder in the bucket
raw_path = './molecular-unfolding-data/117_ideal.mol2' # the mol2 file for this experiment
mol_data = MoleculeData(raw_path, 'qmu')
data_path = mol_data.save("latest")
num_rotation_bond = mol_data.bond_graph.rb_num
print(f"You have loaded the raw molecule data and saved as {data_path}. \n\
This molecule has {num_rotation_bond} rotable bond")
INFO:root:parse mol2 file! INFO:root:finish save qmu_117_ideal_data_latest.pickle
You have loaded the raw molecule data and saved as ./qmu_117_ideal_data_latest.pickle. This molecule has 23 rotable bond
After running this block, the processed data will be saved as qmu_117_ideal_data_latest.pickle and data_path will be updated. We can see that this molecule has 23 rotatable bonds.
Step 2: Build Model¶
In this part, we build the Quadratic Unconstrained Binary Optimization (QUBO) model for molecular unfolding.
First, we set the following parameters and initialize the QMUQUBO object.
We use the 'pre-calc' method to build the model. This molecule has 23 rotatable bonds and we only test 2 of them, so we set the M to 2. And we want the angle to become $45^o$, so we set the D to 8 (i.e., $8=360^o/45^o$). The A and hubo_qubo_val are test from experiments.
# initial the QMUQUBO object
init_param = {}
method = ['pre-calc']
for mt in method:
if mt == 'pre-calc':
init_param[mt] = {}
init_param[mt]['param'] = ['M', 'D', 'A', 'hubo_qubo_val']
qmu_qubo = QMUQUBO(mol_data, method, **init_param)
INFO:root:initial pre-calculate for constructing molecule QUBO
# set the parameters for model
model_param = {}
# parameters
num_rotation_bond = mol_data.bond_graph.rb_num
method = 'pre-calc'
model_param[method] = {}
# model_param[method]['M'] = range(1, num_rotation_bond+1)
model_param[method]['M'] = [1,2,3]
model_param[method]['D'] = [8]
model_param[method]['A'] = [300]
model_param[method]['hubo_qubo_val'] = [200]
qmu_qubo.build_model(**model_param)
INFO:root:Construct model for M:1,D:8,A:300,hubo_qubo_val:200 6.385644276936849e-05 min INFO:root:Construct model for M:2,D:8,A:300,hubo_qubo_val:200 0.0006583054860432942 min INFO:root:Construct model for M:3,D:8,A:300,hubo_qubo_val:200 0.006797945499420166 min
0
We can use the following method to check the properties of model. This way, we can build many models conveniently. After that, we save the model and update the value of model_path.
# describe the model parameters
model_info = qmu_qubo.describe_model()
INFO:root:method: pre-calc
INFO:root:The model_name should be {M}_{D}_{A}_{hubo_qubo_val}
INFO:root:param: M, value {1, 2, 3}
INFO:root:param: D, value {8}
INFO:root:param: A, value {300}
INFO:root:param: hubo_qubo_val, value {200}
# save the model
model_path = qmu_qubo.save("latest")
print(f"You have built the QUBO model and saved it as {model_path}")
INFO:root:finish save qmu_117_ideal_model_latest.pickle
You have built the QUBO model and saved it as ./qmu_117_ideal_model_latest.pickle
Step 3: Optimize Configuration¶
In this part, we use SA and QA to find the optimized configuration of molecular unfolding. At first, we load the model file using QMUQUBO object
qmu_qubo_optimize = QMUQUBO.load(model_path)
model_info = qmu_qubo_optimize.describe_model()
INFO:root:method: pre-calc
INFO:root:The model_name should be {M}_{D}_{A}_{hubo_qubo_val}
INFO:root:param: M, value {1, 2, 3}
INFO:root:param: D, value {8}
INFO:root:param: A, value {300}
INFO:root:param: hubo_qubo_val, value {200}
We can see the parameters of this model, with M equaling 1,2 or 3, D equaling 8, A equaling 300 and hubo_qubo_val equaling 200. Actually, we can contain multiple models in this file just by giving multiple values for one parameter when creating models.
Actually, we can contain multiple models in this file just by giving multiple values for one parameter when creating models. Then, we need use model_name to get the model for experiments.
# get the model you want to optimize
M = 2
D = 8
A = 300
hubo_qubo_val = 200
model_name = "{}_{}_{}_{}".format(M, D, A, hubo_qubo_val)
method = "pre-calc"
qubo_model = qmu_qubo_optimize.get_model(method, model_name)
We can see that we want to carry out experiment with the QUBO model with M equaling 2. After that, we set the parameters for optimization.
| Parameter | Description | Value |
|---|---|---|
| method | annealing method for QUBO problem | 'dwave-sa': use the simulated annealer in ocean toolkit 'dwave-qa': use the quantum annealer |
| shots | number of reads, refer to dwave-sa and dwave-qa for details | 1 to 10,000 |
| bucket | the s3 bucket to store your results | - |
| prefix | the name of the folder in your s3 bucket | - |
| device | the arn name to run your quantum annealing | 'arn:aws:braket:::device/qpu/d-wave/Advantage_system4' 'arn:aws:braket:::device/qpu/d-wave/DW_2000Q_6' |
Then, we can run the SA for this problem:
method = 'neal-sa'
optimizer_param = {}
optimizer_param['shots'] = 1000
sa_optimizer = Annealer(qubo_model, method, **optimizer_param)
INFO:root:use neal simulated annealer (c++) from dimod
sa_optimize_result = sa_optimizer.fit()
INFO:root:fit() ... INFO:root:neal-sa save to local INFO:root:finish save neal-sa_result.pickle
We can tell that we set the number of shots for SA to 1000. The result is saved as the local file ./sa_result.pickle. Alternatively, we can use QA to solve this problem:
# # dwave device has been moved to marketplace, please refer to the following link
# # https://aws.amazon.com/blogs/quantum-computing/using-d-wave-leap-from-the-aws-marketplace-with-amazon-braket-notebooks-and-braket-sdk/
# method = 'dwave-qa'
# optimizer_param = {}
# optimizer_param['shots'] = 1000
# optimizer_param['bucket'] = s3_bucket # the name of the bucket
# optimizer_param['prefix'] = prefix # the name of the folder in the bucket
# optimizer_param['device'] = "arn:aws:braket:::device/qpu/d-wave/Advantage_system4"
# optimizer_param["embed_method"] = "default"
# qa_optimizer = Annealer(qubo_model, method, **optimizer_param)
# # not create annealing task, only embedding logic
# qa_optimizer.embed()
# # create annealing task
# qa_optimize_result = qa_optimizer.fit()
# # not create annealing task, only embedding logic
# qa_optimizer.embed()
# # create annealing task
# qa_optimize_result = qa_optimizer.fit()
Finally, we can compare the execution time between SA and QA :
print(f"dwave-sa run time {sa_optimize_result['time']}")
dwave-sa run time 0.3223903179168701
We sometimes get the best result that occurs only once. This does not always indicate an error. It is actually the characteristic of the problem or how the problem is formulated. Because we have different linear and quadratic terms that vary by many orders of magnitude. If we set change value of A to some smaller number, like 10 or 100, more occurrences of the best answer will be observed. However, these answers usually break the constraints. For more information about this phenomenon, please refer to this Link.
Step 4: PostProcess Result¶
In this part, we post process the optimizing results for evaluation and visualization. At first, we prepare the following parameters:
| Parameter | Description | Value |
|---|---|---|
| method | annealing method for QUBO problem | 'dwave-sa': use the simulated annealer in ocean toolkit 'dwave-qa': use the quantum annealer |
| raw_path | the path for the original molecule file | './molecule-data/117_ideal.mol2' in this example |
| data_path | the path for the processed molecule file | './qmu_117_ideal_data_latest.mol2' in this example |
| bucket | the s3 bucket to store your results | - |
| prefix | the name of the folder in your s3 bucket | - |
| task_id | the id for your quantum annealing task | '2b5a3b05-1a0e-443a-852c-4ec422a10e59' in this example |
Then we can run the post-process using ResultParser object for SA:
method = "neal-sa"
sa_param = {}
sa_param["raw_path"] = raw_path
sa_param["data_path"] = data_path
sa_process_result = ResultParser(method, **sa_param)
# print(f"{method} result is {sa_process_result.get_all_result()}")
local_time, _ , _, _= sa_process_result.get_time()
print(f"time for {method}: \n \
local time is {local_time}")
INFO:root:_load_raw_result INFO:root:load simulated annealer neal-sa raw result INFO:root:MoleculeData.load() INFO:root:init mol data for final position INFO:root:init mol data for raw position INFO:root:_parse_model_info INFO:root:_init_parameters INFO:root:parse simulated annealer result INFO:root:sa only has local_time!
time for neal-sa:
local time is 0.3223903179168701
sa_atom_pos_data = sa_process_result.generate_optimize_pts()
# save unfold file for visualization and parameters for experiment: 1. volume value 2. relative improvement
sa_result_filepath, sa_result_json = sa_process_result.save_mol_file(f"{timestamp}")
print(f"result path is {sa_result_filepath}, and result optimization file path is {sa_result_json}")
INFO:root:generate_optimize_pts()
INFO:root:var_dict_raw {'15': ['8'], '14': ['1']} var_dict_list [{'15': '8', '14': '1'}]
INFO:root:chosen var {'x_15_8', 'x_14_1'}
INFO:root:tor list {'X_15_8', 'X_14_1'}
INFO:root:initial 18.549876807564768
INFO:root:optimize 18.561854117306133
INFO:root:optimize_gain 1.0006456813630418
INFO:root:start physical check
INFO:root:save_mol_file 20230527-12
INFO:root:finish save ./molecular-unfolding-data/117_ideal_neal-sa_20230527-12.mol2 and ./molecular-unfolding-data/117_ideal_neal-sa_20230527-12.json
result path is ./molecular-unfolding-data/117_ideal_neal-sa_20230527-12.mol2, and result optimization file path is ./molecular-unfolding-data/117_ideal_neal-sa_20230527-12.json
sa_process_result.parameters
{'volume': {'optimize': 18.561854117306133,
'initial': 18.549876807564768,
'gain': 1.0006456813630418,
'unfolding_results': ['X_15_8', 'X_14_1'],
'annealing_results': ['x_15_8', 'x_14_1'],
'optimize_info': {'optimize_state': True, 'result_rank': 2}}}
In the first block, we can see the local time for SA is more than 150 seconds. With the generate_optimize_pts() method, the final 3D points after unfolding will be generated and saved as json file and mol2 files. The last block shows the optimizing results which are also stored in json files. It shows that the optimized result gains 1.0006x increase in volume. The value for unfolding_results indicates that the rotatable bond 15 should rotate $270^o$ ($360/8*(7-1)$) and the rotatable bond 14 should rotate $0^o$ ($360/8*(1-1)$). At the same time, you can run the post-process for QA:
# The following codes can be used as the reference once you can access d-wave devices from marketplace
# method = "dwave-qa"
# qa_param = {}
# qa_param["bucket"] = s3_bucket
# qa_param["prefix"] = prefix
# qa_param["task_id"] = qa_task_id
# qa_param["raw_path"] = raw_path
# qa_param["data_path"] = data_path
# qa_process_result = ResultParser(method, **qa_param)
# # print(f"{method} result is {qa_process_result.get_all_result()}")
# local_time, task_time, total_time, access_time = qa_process_result.get_time()
# print(f"time for {method}: \n \
# local time is {local_time},\n \
# task time is {task_time}, \n \
# qpu total time is {total_time}, \n \
# qpu access time is {access_time}")
we can see that there many types of time metrics for running QA. This task has the local time of 5 s, which means the time between calling the api and getting the annealing result. The task time time is the metric from the json file in bucket. We can also see the qpu total time and qpu access time representing the actual time running in the QPU. Please refer to Operation and Timing for details.
# qa_atom_pos_data = qa_process_result.generate_optimize_pts()
# # save unfold file for visualization and parameters for experiment: 1. volume value 2. relative improvement
# qa_result_filepath, qa_result_json = qa_process_result.save_mol_file(f"{timestamp}")
# print(f"result path is {qa_result_filepath}, and result optimization file path is {qa_result_json}")
# qa_process_result.parameters
Finally, We can open folders for the optimized results:
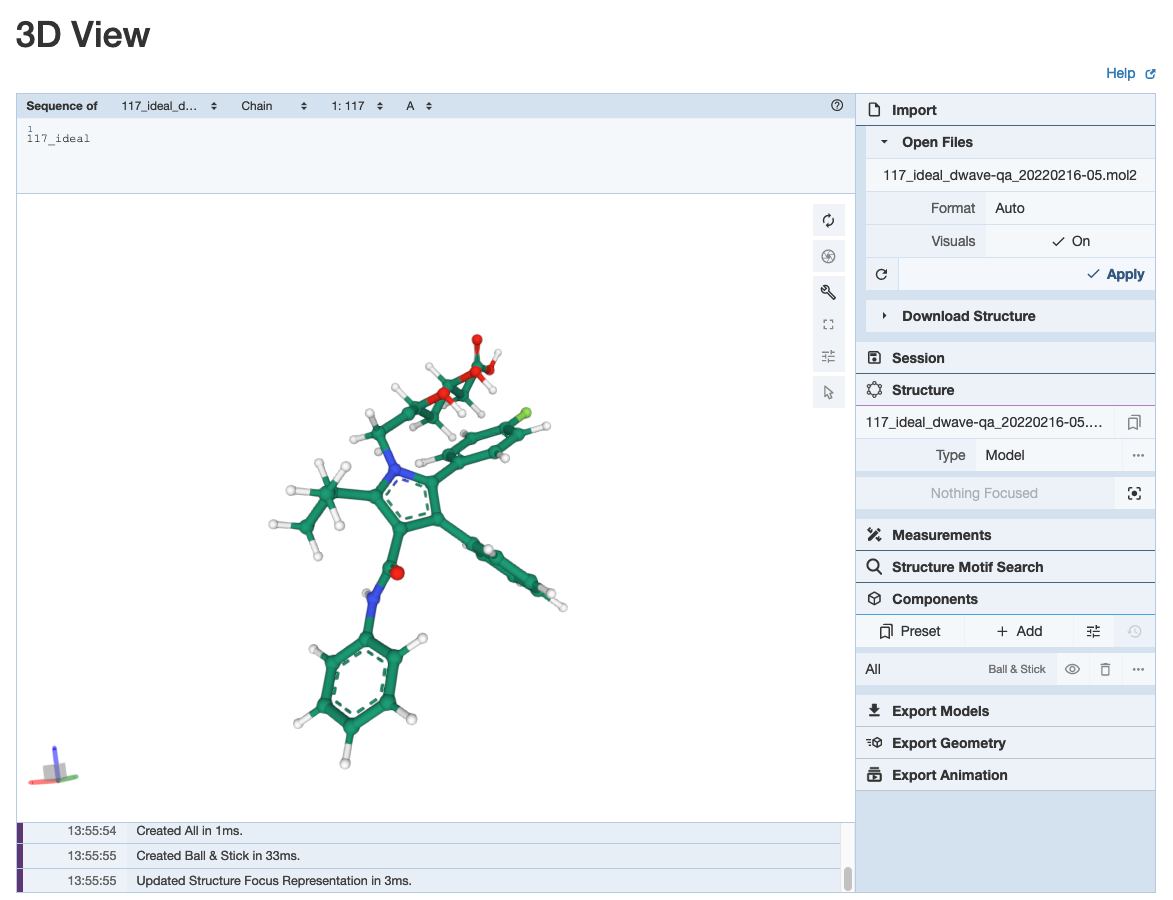
We can see the json result and mol2 file of SA and QA are stored in this place. If we carry out more experiments, more results with time stamp are stored incrementally. For visualization, we can upload the mol2 file into online viewer tool to see the result:
Hybrid Job Experiment¶
from braket.aws import AwsQuantumJob
from braket.jobs.config import InstanceConfig
import boto3
import json
import altair as alt
import pandas as pd
import numpy as np
from hybridjobs.utility.HybridJobHelpers import *
Step 1: Prepare parameters for batch evaluation¶
In this part, we set the parameters for batch evaluation
# parameters for experiments
experiment_name = "molecular-unfolding-qubo"
data_path = "molecular-unfolding-data"
experiments_params = {
"version": "1",
"params": [
{"M": [1,2,3,4,5,6]},
{"D": [8]},
{"shots": [10000]},
{"device": [{"qc": "null", "cc": "ml.m5.large"},{"qc": "null", "cc": "ml.m5.4xlarge"}]}
]
}
hp = {}
hybrid_job_params = []
parse_params(experiments_params['params'], hp, hybrid_job_params)
print(f"parameters for experiments: \n {hybrid_job_params}")
parameters for experiments:
[{'M': 1, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 1, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}, {'M': 2, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 2, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}, {'M': 3, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 3, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}, {'M': 4, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 4, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}, {'M': 5, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 5, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}, {'M': 6, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.large'}}, {'M': 6, 'D': 8, 'shots': 10000, 'device': {'qc': 'null', 'cc': 'ml.m5.4xlarge'}}]
# Upload dataset to S3
s3_path = upload_data(data_path)
print(f"upload data to s3 path: {s3_path}")
upload data to s3 path: s3://amazon-braket-us-east-1-002224604296/molecular-unfolding-data
Step 2: Prepare image for experiment¶
In this part, we use the following code to prepare the image for experiment. For the first run, please run build_and_push.sh to create the image. For future experiments, avoid running build_and_push.sh unless you want to rebuild the image
account_id = boto3.client("sts").get_caller_identity()["Account"]
region = boto3.client('s3').meta.region_name
image_name = f"amazon-braket-{experiment_name}-jobs"
image_uri = f"{account_id}.dkr.ecr.{region}.amazonaws.com/{image_name}:latest"
print(f"the hybrid job image for {account_id} in region {region}: {image_uri}")
!sh build_and_push.sh {image_name}
the hybrid job image for 002224604296 in region us-east-1: 002224604296.dkr.ecr.us-east-1.amazonaws.com/amazon-braket-molecular-unfolding-jobs:latest WARNING! Your password will be stored unencrypted in /home/ubuntu/.docker/config.json. Configure a credential helper to remove this warning. See https://docs.docker.com/engine/reference/commandline/login/#credentials-store Login Succeeded WARNING! Your password will be stored unencrypted in /home/ubuntu/.docker/config.json. Configure a credential helper to remove this warning. See https://docs.docker.com/engine/reference/commandline/login/#credentials-store Login Succeeded Sending build context to Docker daemon 1.224MB Step 1/3 : FROM 292282985366.dkr.ecr.us-west-2.amazonaws.com/amazon-braket-base-jobs:1.0-cpu-py37-ubuntu18.04 ---> 16b9ec942e00 Step 2/3 : RUN python3 -m pip install --upgrade pip ---> Using cache ---> 11a512e96ae1 Step 3/3 : RUN python3 -m pip install dimod==0.10.12 dwave-system==1.15.0 dwave-neal==0.5.9 networkx==2.6.3 amazon-braket-ocean-plugin==1.0.7 Cython==0.29.32 biopandas==0.4.1 py3Dmol==1.8.0 ipywidgets==7.7.0 ---> Using cache ---> ee4738be5da2 Successfully built ee4738be5da2 Successfully tagged amazon-braket-molecular-unfolding-jobs:latest The push refers to repository [002224604296.dkr.ecr.us-east-1.amazonaws.com/amazon-braket-molecular-unfolding-jobs] 367c4a78: Preparing b90ba34e: Preparing 6411a7d1: Preparing 56d89649: Preparing 148e4f08: Preparing 0f2f3775: Preparing a5237a47: Preparing 7b584848: Preparing 9cc6ed2d: Preparing 9ba03cb5: Preparing 3dae4964: Preparing 0cee8562: Preparing c13d298e: Preparing 2ecf6ff1: Preparing 5cb74c43: Preparing 49c05c79: Preparing b7118beb: Preparing 5cb74c43: Layer already exists latest: digest: sha256:fab303fac8662a402c81beda3c2421325345033537eb47fce3f6d55c82588d08 size: 4104
hybrid_jobs_json = f"{experiment_name}-hybrid-jobs.json"
print(f"job info will be saved in {hybrid_jobs_json}")
job info will be saved in molecular-unfolding-qubo-hybrid-jobs.json
Step 3: Launch Amazon Braket Hybrid Jobs for experiment¶
In this part, we use the following code to launch the same number of hybrid jobs as the sets of parameters for this experiments. When the number of jobs exceeds 5, this thread will wait. The default setting of this experiment will take less than 1 hour to finish.
# Long runnning cell due to Burst rate of CreateJob requests < 5 RPS
# sudo apt-get install python-prctl at first
# https://stackoverflow.com/questions/34361035/python-thread-name-doesnt-show-up-on-ps-or-htop
from threading import Thread
import threading
import setproctitle
import uuid
U=str(uuid.uuid4())[:4]
def launch_hybrid_jobs(hybrid_job_params=hybrid_job_params, hybrid_jobs_json=hybrid_jobs_json):
setproctitle.setproctitle(threading.current_thread().name)
# parse evaluation parameters and trigger hybrid jobs:
jobs = []
names = []
job_name = f"{experiment_name}-job"
job_count = len(hybrid_job_params)
# from braket.jobs.local import LocalQuantumJob
for job_param in hybrid_job_params:
M = job_param['M']
D = job_param['D']
quantum_device = get_quantum_device(job_param['device']['qc'])
classical_device = job_param['device']['cc']
device_name = classical_device.replace(".","-")
device_name = device_name.replace("x","")
job_param['job_count'] = job_count
name = f"{U}-{experiment_name}-M-{M}-D-{D}-{device_name}"
tmp_job = AwsQuantumJob.create(
device=quantum_device,
source_module="hybridjobs",
entry_point=f"hybridjobs.{job_name}:main",
job_name=name,
hyperparameters=job_param,
input_data=s3_path,
instance_config=InstanceConfig(instanceType=classical_device),
image_uri=image_uri,
wait_until_complete=False,
)
print(f"Finish create {experiment_name} with M {M}, D {D} and device {device_name}")
jobs.append(tmp_job)
names.append(name)
while not queue_check(jobs):
time.sleep(5)
jobs_arn = []
for job in jobs:
jobs_arn.append(job.arn)
jobs_states = {
"experiment_name": experiment_name,
"hybrid-jobs-arn": jobs_arn,
"names": names
}
# save hybrid job arn for further analysis
json_object = json.dumps(jobs_states, indent=4)
with open(hybrid_jobs_json, "w") as outfile:
outfile.write(json_object)
print(f"Finish launch all the hybrid jobs and save all the files")
t = Thread(target=launch_hybrid_jobs, name="launch-hybrid-job", daemon=True).start()
INFO:botocore.credentials:Found credentials in shared credentials file: ~/.aws/credentials INFO:botocore.credentials:Found credentials in shared credentials file: ~/.aws/credentials INFO:botocore.credentials:Found credentials in shared credentials file: ~/.aws/credentials INFO:botocore.credentials:Found credentials in shared credentials file: ~/.aws/credentials INFO:botocore.credentials:Found credentials in shared credentials file: ~/.aws/credentials
fail to get null: list index out of range, use sv1 instead Finish create molecular-unfolding with M 1, D 8 and device ml-m5-large
# run the following scripts to check the created threads
!ps -aux | grep launch-hybrid-job
ubuntu 34835 1.3 1.5 1865832 487616 ? Sl 12:01 0:47 launch-hybrid-job ubuntu 37348 0.0 0.0 8756 3712 pts/1 Ss+ 12:57 0:00 /bin/bash -c ps -aux | grep launch-hybrid-job ubuntu 37353 0.0 0.0 8176 720 pts/1 S+ 12:57 0:00 grep launch-hybrid-job There are 4 jobs in RUNNING or QUEUED status There are 3 jobs in RUNNING or QUEUED status Finish launch all the hybrid jobs and save all the files
Step 4: Jobs finish and visualize results¶
Please use the following code to check the status of hybrid jobs. The status of hybrid jobs can also be checked in the Amazon Braket console. Optionally, if the email if input when deploying the solution, emails will be sent at the same number of hybrid jobs once the status of jobs changes.
# run the following code to test whether all the jobs finish
results = []
if os.path.exists(hybrid_jobs_json):
# recover hybrid jobs and show result
jobs_states_load = None
with open(hybrid_jobs_json, "r") as outfile:
jobs_states_load = json.load(outfile)
completed_jobs_arn = set()
for job_name, job_arn in zip(jobs_states_load["names"], jobs_states_load["hybrid-jobs-arn"]):
current_job = AwsQuantumJob(job_arn)
print(f"the state of job {job_name} is : {current_job.state()}")
if current_job.state() == 'COMPLETED':
completed_jobs_arn.update({job_arn})
whole_jobs_num = len(jobs_states_load["names"])
results = None
if len(completed_jobs_arn) == whole_jobs_num:
print(f"all jobs completed")
results = []
for job_arn in completed_jobs_arn:
current_job = AwsQuantumJob(job_arn)
results.append(current_job.result())
print(current_job.result())
# display results
results = display_results(results, experiments_params)
else:
print(f"JSON file for job arns not generated! please wait for the thread(launch-hybrid-job) to finish")
the state of job f3f2-molecular-unfolding-M-1-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-1-D-8-ml-m5-4large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-2-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-2-D-8-ml-m5-4large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-3-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-3-D-8-ml-m5-4large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-4-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-4-D-8-ml-m5-4large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-5-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-5-D-8-ml-m5-4large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-6-D-8-ml-m5-large is : COMPLETED
the state of job f3f2-molecular-unfolding-M-6-D-8-ml-m5-4large is : COMPLETED
all jobs completed
{'time': 320.81806111335754, 'hypermeter': {'D': '8', 'M': '5', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 94.67504777232924, 'initial': 94.67504777232924, 'gain': 1.0, 'unfolding_results': ['X_12_1', 'X_15_1', 'X_14_1', 'X_11_1', 'X_18_1'], 'annealing_results': ['X_12_1', 'X_15_1', 'X_14_1', 'X_11_1', 'X_18_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 32}}}}
{'time': 61.88261556625366, 'hypermeter': {'D': '8', 'M': '4', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 65.63612176874935, 'initial': 65.63612176874935, 'gain': 1.0, 'unfolding_results': ['X_12_1', 'X_14_1', 'X_15_1', 'X_18_1'], 'annealing_results': ['X_12_1', 'X_14_1', 'X_15_1', 'X_18_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 1335}}}}
{'time': 20.689651012420654, 'hypermeter': {'D': '8', 'M': '3', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 32.05739524950204, 'initial': 32.05739524950204, 'gain': 1.0, 'unfolding_results': ['X_12_1', 'X_14_1', 'X_15_1'], 'annealing_results': ['X_12_1', 'X_14_1', 'X_15_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 39}}}}
{'time': 2.633816957473755, 'hypermeter': {'D': '8', 'M': '1', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 8.11248514215326, 'initial': 8.11248514215326, 'gain': 1.0, 'unfolding_results': ['X_15_1'], 'annealing_results': ['X_15_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 2}}}}
{'time': 3.6196465492248535, 'hypermeter': {'D': '8', 'M': '2', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 18.561870734607446, 'initial': 18.561870734607446, 'gain': 1.0, 'unfolding_results': ['X_15_1', 'X_14_1'], 'annealing_results': ['X_15_1', 'X_14_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 7}}}}
{'time': 2.5614542961120605, 'hypermeter': {'D': '8', 'M': '1', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 8.11248514215326, 'initial': 8.11248514215326, 'gain': 1.0, 'unfolding_results': ['X_15_1'], 'annealing_results': ['X_15_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 9}}}}
{'time': 3.6330432891845703, 'hypermeter': {'D': '8', 'M': '2', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 18.561870734607446, 'initial': 18.561870734607446, 'gain': 1.0, 'unfolding_results': ['X_15_1', 'X_14_1'], 'annealing_results': ['X_15_1', 'X_14_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 17}}}}
{'time': 568.0857925415039, 'hypermeter': {'D': '8', 'M': '6', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 134.28621271340106, 'initial': 134.28621271340106, 'gain': 1.0, 'unfolding_results': ['X_12_1', 'X_11_1', 'X_18_1', 'X_15_1', 'X_14_1', 'X_21_1'], 'annealing_results': ['X_12_1', 'X_11_1', 'X_18_1', 'X_15_1', 'X_14_1', 'X_21_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 6691}}}}
{'time': 21.914642095565796, 'hypermeter': {'D': '8', 'M': '3', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 32.05739524950203, 'initial': 32.05739524950203, 'gain': 1.0, 'unfolding_results': ['X_14_1', 'X_15_1', 'X_12_1'], 'annealing_results': ['X_14_1', 'X_15_1', 'X_12_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 413}}}}
{'time': 315.054883480072, 'hypermeter': {'D': '8', 'M': '5', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 94.17211104523905, 'initial': 94.17211104523905, 'gain': 1.0, 'unfolding_results': ['X_15_1', 'X_18_1', 'X_11_1', 'X_14_1', 'X_12_1'], 'annealing_results': ['X_15_1', 'X_18_1', 'X_11_1', 'X_14_1', 'X_12_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 31}}}}
{'time': 64.51841259002686, 'hypermeter': {'D': '8', 'M': '4', 'device': "{'qc': 'null', 'cc': 'ml.m5.large'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 65.63612176874935, 'initial': 65.63612176874935, 'gain': 1.0, 'unfolding_results': ['X_15_1', 'X_14_1', 'X_18_1', 'X_12_1'], 'annealing_results': ['X_15_1', 'X_14_1', 'X_18_1', 'X_12_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 482}}}}
{'time': 553.271329164505, 'hypermeter': {'D': '8', 'M': '6', 'device': "{'qc': 'null', 'cc': 'ml.m5.4xlarge'}", 'job_count': '12', 'shots': '10000'}, 'result': {'volume': {'optimize': 134.28621271340106, 'initial': 134.28621271340106, 'gain': 1.0, 'unfolding_results': ['X_18_1', 'X_21_1', 'X_12_1', 'X_15_1', 'X_14_1', 'X_11_1'], 'annealing_results': ['X_18_1', 'X_21_1', 'X_12_1', 'X_15_1', 'X_14_1', 'X_11_1'], 'optimize_info': {'optimize_state': False, 'result_rank': 1827}}}}
rename_result = {}
device_list = []
x_list = []
y_list = []
for k,vs in results.items():
k = k.replace("\'","\"")
dict_k = json.loads(k)
device_name = None
if dict_k['qc'] == 'null':
device_name = dict_k['cc']
else:
device_name = dict_k['qc']
for v in vs:
device_list.append(device_name)
x_list.append(v[2])
y_list.append(v[3])
source = pd.DataFrame({
"DxM": np.array(x_list),
"Time to Solution": np.array(y_list),
"Device": np.array(device_list),
})
alt.Chart(source).mark_line(point = True).encode(
x='DxM',
y='Time to Solution',
color='Device',
).properties(
title = "Molecular unfolding experiments",
width = 700,
height = 600,
).interactive()