Notebook experimentation
This notebook implements the protein folding using varionational quantum eigensolver (vqe).
Protein folding using VQE
Protein folding is the process by which a protein molecule assumes its three-dimensional shape, which is essential for its proper function. Proteins are made up of a linear chain of amino acids, and their final structure is determined by the sequence of amino acids and the interactions between them.
During protein folding, the linear chain of amino acids folds into a unique three-dimensional structure, which is stabilized by various types of interactions, such as hydrogen bonds, electrostatic forces, and van der Waals forces. The process of protein folding is highly complex and involves multiple stages, including the formation of secondary structures, such as alpha-helices and beta-sheets, and the packing of these structures into a final three-dimensional shape.
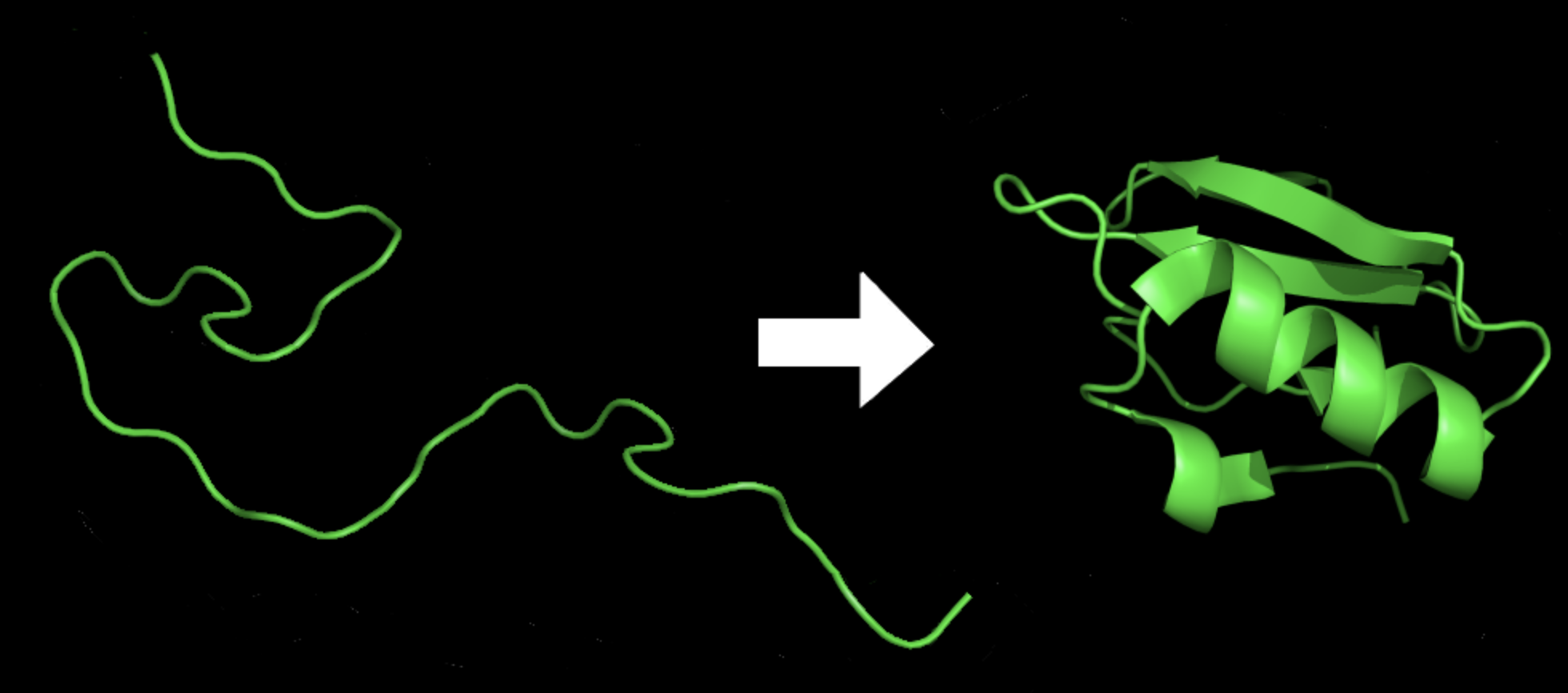
Figure 10: Protein before and after folding10
VQE is a quantum algorithm that can be used to simulate the behavior of molecules, including proteins. VQE works by approximating the ground state energy of a molecule using a quantum computer, which can be used to predict the properties of the molecule.
To use VQE for protein folding, one would first need to encode the protein structure into a format that can be represented as a quantum state. This can be done using a technique called the qubitization method, which maps the protein structure onto a set of qubits that can be manipulated by the quantum computer.
After deployment, you can go to select the solution’s root stack on the Stacks page, choose the Outputs tab, and open the link for your notebook. See the file healthcare-and-life-sciences/c-2-protein-folding-variational-quantum-eigensolver/protein-folding-vqe.ipynb for more details.